| Home | Sources Directory | News Releases | Calendar | Articles | | Contact | |
Transfer RNA
Transfer RNA (tRNA) is a small RNA molecule (usually about 74-95 nucleotides) that transfers a specific active amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis during translation. It has a 3' terminal site for amino acid attachment. This covalent linkage is catalyzed by an aminoacyl tRNA synthetase. It also contains a three base region called the anticodon that can base pair to the corresponding three base codon region on mRNA. Each type of tRNA molecule can be attached to only one type of amino acid, but because the genetic code contains multiple codons that specify the same amino acid, tRNA molecules bearing different anticodons may also carry the same amino acid.
Contents |
[edit] Structure
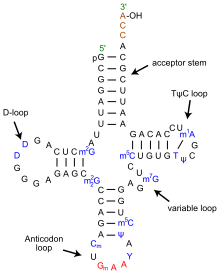
The structure of tRNA can be decomposed into its primary structure, its secondary structure (usually visualized as the cloverleaf structure), and its tertiary structure (all tRNAs have a similar L-shaped 3D structure that allows them to fit into the P and A sites of the ribosome). The cloverleaf structure becomes the 3D L-shaped structure through coaxial stacking of the helices which is a common RNA Tertiary Structure motif.
- The 5'-terminal phosphate group.
- The acceptor stem is a 7-base pair (bp) stem made by the base pairing of the 5'-terminal nucleotide with the 3'-terminal nucleotide (which contains the CCA 3'-terminal group used to attach the amino acid). The acceptor stem may contain non-Watson-Crick base pairs.
- The CCA tail is a cytosine-cytosine-adenine sequence at the 3' end of the tRNA molecule. This sequence is important for the recognition of tRNA by enzymes critical in translation. In prokaryotes, the CCA sequence is transcribed in some tRNA sequences. In most prokaryotic tRNAs and eukaryotic tRNAs, the CCA sequence is added during processing and therefore does not appear in the tRNA gene.
- The D arm is a 4 bp stem ending in a loop that often contains dihydrouridine.
- The anticodon arm is a 5-bp stem whose loop contains the anticodon.
- The T arm is a 5 bp stem containing the sequence Tî�C where î� is a pseudouridine.
- Bases that have been modified, especially by methylation, occur in several positions outside the anticodon. The first anticodon base is sometimes modified to inosine (derived from adenine) or pseudouridine (derived from uracil).
[edit] Anticodon
An anticodon[1] is a unit made up of three nucleotides that correspond to the three bases of the codon on the mRNA. Each tRNA contains a specific anticodon triplet sequence that can base-pair to one or more codons for an amino acid. For example, the codon for lysine is AAA; the anticodon of a lysine tRNA might be UUU. Some anticodons can pair with more than one codon due to a phenomenon known as wobble base pairing. Frequently, the first nucleotide of the anticodon is one of two not found on mRNA: inosine and pseudouridine, which can hydrogen bond to more than one base in the corresponding codon position. In the genetic code, it is common for a single amino acid to be specified by all four third-position possibilities, or at least by both Pyrimidines and Purines; for example, the amino acid glycine is coded for by the codon sequences GGU, GGC, GGA, and GGG.
To provide a one-to-one correspondence between tRNA molecules and codons that specify amino acids, 61 types of tRNA molecules would be required per cell. However, many cells contain fewer than 61 types of tRNAs because the wobble base is capable of binding to several, though not necessarily all, of the codons that specify a particular amino acid. A minimum of 31 tRNA are required to translate, unambiguously, all 61 sense codons of the standard genetic code.[2]
[edit] Aminoacylation
Aminoacylation is the process of adding an aminoacyl group to a compound. It produces tRNA molecules with their CCA 3' ends covalently linked to an amino acid.
Each tRNA is aminoacylated (or charged) with a specific amino acid by an aminoacyl tRNA synthetase. There is normally a single aminoacyl tRNA synthetase for each amino acid, despite the fact that there can be more than one tRNA, and more than one anticodon, for an amino acid. Recognition of the appropriate tRNA by the synthetases is not mediated solely by the anticodon, and the acceptor stem often plays a prominent role.
Reaction:
Sometimes, certain organisms can have one or more aminoacyl tRNA synthetases missing. This leads to mischarging of the tRNA by a chemically related amino acid. The correct amino acid is made by enzymes that modify the mischarged amino acid to the correct one.
For example, Helicobacter pylori has glutaminyl tRNA synthetase missing. Thus, glutamate tRNA synthetase mischarges tRNA-glutamine(tRNA-Gln) with glutamate. An amidotransferase then converts the acid side chain of the glutamate to the amide, forming the correctly charged gln-tRNA-Gln.
[edit] Binding to ribosome
The ribosome has three binding sites for tRNA molecules: the A (aminoacyl), P (peptidyl), and E (exit) sites. During translation the A site binds an incoming aminoacyl-tRNA as directed by the codon currently occupying this site. This codon specifies the next amino acid to be added to the growing peptide chain. The A site only works after the first aminoacyl-tRNA has attached to the P site. The P-site codon is occupied by peptidyl-tRNA that is a tRNA with multiple amino acids attached as a long chain. The P site is actually the first to bind to aminoacyl tRNA. This tRNA in the P site carries the chain of amino acids that has already been synthesized. The E site is occupied by the empty tRNA as it's about to exit the ribosome.
[edit] tRNA genes
Organisms vary in the number of tRNA genes in their genome. The nematode worm C. elegans, a commonly used model organism in genetics studies, has 29,647 [3] genes in its nuclear genome, of which 620 code for tRNA.[4][5] The budding yeast Saccharomyces cerevisiae has 275 tRNA genes in its genome. In the human genome, which according to current estimates has about 27,161 genes [6] in total, there are about 4,421 non-coding RNA genes, which include tRNA genes. There are 22 mitochondrial tRNA genes;[7] 497 nuclear genes encoding cytoplasmic tRNA molecules and there are 324 tRNA-derived putative pseudogenes.[8]
Cytoplasmic tRNA genes can be grouped into 49 families according to their anticodon features. These genes are found on all chromosomes, except 22 and Y chromosome. High clustering on 6p is observed (140 tRNA genes), as well on 1 chromosome.[8]
[edit] tRNA biogenesis
In eukaryotic cells, tRNAs are transcribed by RNA polymerase III as pre-tRNAs in the nucleus.[9] RNA polymerase III recognizes two internal promoter sequences (A-box B internal promoter) inside tRNA genes.[10] The first promoter begins at nucleotide 8 of mature tRNAs and the second promoter is located 30-60 nucleotides downstream of the first promoter. The transcription terminates after a strech of four or more thymidines.[10]
Pre-tRNAs undergo extensive modifications inside the nucleus. Some pre-tRNAs contain introns;[citation needed] in bacteria these self-splice, whereas in eukaryotes and archaea they are removed by tRNA splicing endonuclease.[11]. The 5' sequence is removed by RNase P,[12] whereas the 3' end is removed by the tRNase Z enzyme.[13] A notable exception is in the archaeon Nanoarchaeum equitans which does not possess an RNase P enzyme and has a promoter placed such that transcription starts at the 5' end of the mature tRNA.[14]. The non-templated 3' CCA tail is added by a nucleotidyl transferase.[15] Before tRNAs are exported into the cytoplasm by Los1/Xpo-t,[16][17] tRNAs are aminoacylated.[18] The order of the processing events is not conserved. For example in yeast, the splicing is not carried out in the nucleus but at the cytoplasmic side of mitochondrial membranes.[19]
[edit] History
The existence of tRNA was first hypothesized by Francis Crick, based on the assumption that there must exist an adapter molecule capable of mediating the translation of the RNA alphabet into the protein alphabet. Significant research on structure was conducted in the early 1960s by Alex Rich and Don Caspar, two researchers in Boston, the Jacques Fresco group in Princeton University and a United Kingdom group at King's College London.[20] In 1965, a publication by Robert W. Holley reported the primary structure and suggested three secondary structures.[21] The cloverleaf structure was ascertained by several other studies in the following years[22] and was finally confirmed using X-ray crystallography studies in 1974. Two independent groups, Kim Sung-Hou working under Alexander Rich and a British group headed by Aaron Klug, published the same crystallography findings within a year.[23][24]
[edit] See also
- mRNA
- tmRNA
- non-coding RNA and introns
- translation
- Transfer RNA-like structures
- Kim Sung-Hou
- wobble hypothesis
[edit] References
- ^ Felsenfeld G, Cantoni G (1964). "Use of thermal denaturation studies to investigate the base sequence of yeast serine sRNA". Proc Natl Acad Sci USA 51: 818'26. doi:10.1073/pnas.51.5.818. PMID 14172997.
- ^ Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky SL, Darnell J. (2004). Molecular Biology of the Cell. WH Freeman: New York, NY. 5th ed.
- ^ WormBase web site, http://www.wormbase.org, release WS187, date 25-Jan-2008.
- ^ Spieth, J; Lawson, D (Jan 2006). "Overview of gene structure." (Free full text). WormBook : the online review of C. Elegans biology: 1'10. doi:10.1895/wormbook.1.65.1. PMID 18023127.
- ^ Hartwell LH, Hood L, Goldberg ML, Reynolds AE, Silver LM, Veres RC. (2004). Genetics: From Genes to Genomes 2nd ed. McGraw-Hill: New York, NY. p 264.
- ^ Ensembl release 48 - Dec 2007 http://www.ensembl.org
- ^ Ibid. p 529.
- ^ a b Lander E. et al. (2001). "Initial sequencing and analysis of the human genome". Nature 409 (6822): 860'921. doi:10.1038/35057062. PMID 11237011.
- ^ White RJ (1997). "Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control?". Trends in Biochemical Sciences 22 (3): 77'80. doi:10.1016/S0968-0004(96)10067-0. PMID 9066256.
- ^ a b Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A (December 2007). "The expanding RNA polymerase III transcriptome". Trends Genet. 23 (12): 614'22. doi:10.1016/j.tig.2007.09.001. PMID 17977614.
- ^ Abelson J, Trotta CR, Li H (1998). "tRNA Splicing". J Biol Chem 273 (21): 12685'12688. doi:10.1074/jbc.273.21.12685. PMID 9582290.
- ^ Frank DN, Pace NR (1998). "Ribonuclease P: unity and diversity in a tRNA processing ribozyme". Annu. Rev. Biochem. 67: 153'80. doi:10.1146/annurev.biochem.67.1.153. PMID 9759486.
- ^ Ceballos M, Vioque A (2007). "tRNase Z". Protein Pept. Lett. 14 (2): 137'45. doi:10.2174/092986607779816050. PMID 17305600.
- ^ Randau L, Schröder I, Söll D (May 2008). "Life without RNase P". Nature 453 (7191): 120'3. doi:10.1038/nature06833. PMID 18451863.
- ^ Weiner AM (October 2004). "tRNA maturation: RNA polymerization without a nucleic acid template". Curr. Biol. 14 (20): R883'5. doi:10.1016/j.cub.2004.09.069. PMID 15498478.
- ^ Kutay, U.; Lipowsky, G.; Izaurralde, E.; Bischoff, F.; Schwarzmaier, P.; Hartmann, E.; Görlich, D. (1998). "Identification of a tRNA-Specific Nuclear Export Receptor". Molecular Cell 1 (3): 359'369. doi:10.1016/S1097-2765(00)80036-2. PMID 9660920.
- ^ Arts, G. J.; Fornerod, M.; Mattaj, L. W. (1998). "Identification of a nuclear export receptor for tRNA". Current Biology 8 (6): 305. doi:10.1016/S0960-9822(98)70130-7. PMID 9512417.
- ^ Arts, G.; Kuersten, S.; Romby, P.; Ehresmann, B.; Mattaj, I. (1998). "The role of exportin-t in selective nuclear export of mature tRNAs". The EMBO journal 17 (24): 7430'7441. doi:10.1093/emboj/17.24.7430. PMID 9857198.
- ^ Yoshihisa, T.; Yunoki-Esaki, K.; Ohshima, C.; Tanaka, N.; Endo, T. (2003). "Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria". Molecular biology of the cell 14 (8): 3266'3279. doi:10.1091/mbc.E02-11-0757. PMID 12925762.
- ^ Brian F.C. Clark (October 2006). "The crystal structure of tRNA". J. Biosci. 31 (4): 453'7. doi:10.1007/BF02705184. PMID 17206065. http://www.ias.ac.in/jbiosci/oct2006/453.pdf.
- ^ HOLLEY RW, APGAR J, EVERETT GA, et al. (March 1965). "STRUCTURE OF A RIBONUCLEIC ACID". Science 147: 1462'5. PMID 14263761. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=14263761. Retrieved 2010-09-03.
- ^ "The Nobel Prize in Physiology or Medicine 1968". Nobel Foundation. http://nobelprize.org/nobel_prizes/medicine/laureates/1968/index.html. Retrieved 2007-07-28.
- ^ Ladner JE, Jack A, Robertus JD, et al. (November 1975). "Structure of yeast phenylalanine transfer RNA at 2.5 A resolution". Proc. Natl. Acad. Sci. U.S.A. 72 (11): 4414'8. PMID 1105583.
- ^ Kim SH, Quigley GJ, Suddath FL, et al. (1973). "Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain". Science 179 (70): 285'8. doi:10.1126/science.179.4070.285. PMID 4566654.
[edit] External links
- tRNAdb (updated and completely restructured version of Spritzls tRNA compilation)
- original Sprinzl tRNA compilation
- tRNA link to heart disease and stroke
- GtRNAdb: Collection of tRNAs identified from complete genomes
- Molecule of the Month – RCSB Protein Data Bank:
|
||||||||||||||||||||
|
SOURCES.COM is an online portal and directory for journalists, news media, researchers and anyone seeking experts, spokespersons, and reliable information resources. Use SOURCES.COM to find experts, media contacts, news releases, background information, scientists, officials, speakers, newsmakers, spokespeople, talk show guests, story ideas, research studies, databases, universities, associations and NGOs, businesses, government spokespeople. Indexing and search applications by Ulli Diemer and Chris DeFreitas.
For information about being included in SOURCES as a expert or spokesperson see the FAQ . For partnerships, content and applications, and domain name opportunities contact us.